A few words on Clades & Cladograms |Taxonomy 5
[By Chuck Almdale]
We previously traveled through the Kingdoms of Protista, Fungi, Plantae and have journeyed most of the way through Kingdom Animalia. We have one brief stop before we enter Phylum Chordata. The above diagram is the nicely-designed cladogram we used for the plant kingdom in Taxonomy 3, complete with dates of divergence and synapomorphies. From: Cal Poly Humboldt Natural History Museum
The above diagram is the nicely-designed cladogram we used for the plant kingdom in Taxonomy 3, complete with dates of divergence and synapomorphies. From: Cal Poly Humboldt Natural History Museum
Cladistics
We have mentioned clades a few times in passing, but as we’re about to plunge deeper into them we need to introduce a few terms and concepts. The term “clade” first appeared almost 70 years ago, but it took a few decades for it to become accepted and widely use. Before the discovery and use of evolutionary clocks, classification and ranking was based on morphology; the concept of connecting species to their proper ancestor, and then speaking of the entire group – one common ancestor and all its descendants whether extant or extinct – was a goal difficult if not impossible to achieve with a high (or perhaps even low) degree of certainty. After the discovery (or creation) of the biological clocks and the beginning of graphic representations of the relationships between organisms – often including elapsed time as one of the axes – scientists quickly realized they were now capable of reaching a far higher level of detail with a high level of reliability. They also saw a problem in taxonomic nomenclature approaching.
The approximately thirty ranks in the Linnaean system were far too few. There were potentially hundreds – perhaps many thousands – of rank levels waiting to be discovered and named. The day could quickly arrive when words would literally fail to describe. They needed a simpler method of depicting the proliferation of newly-discovered relationships between organisms and their ancestral lineages, and the cladogram was the result. With the new method of description and presentation, a new terminology had to be created. The following definitions are like an evolutionary tree, in that the fundamentals are presented before the specifics.
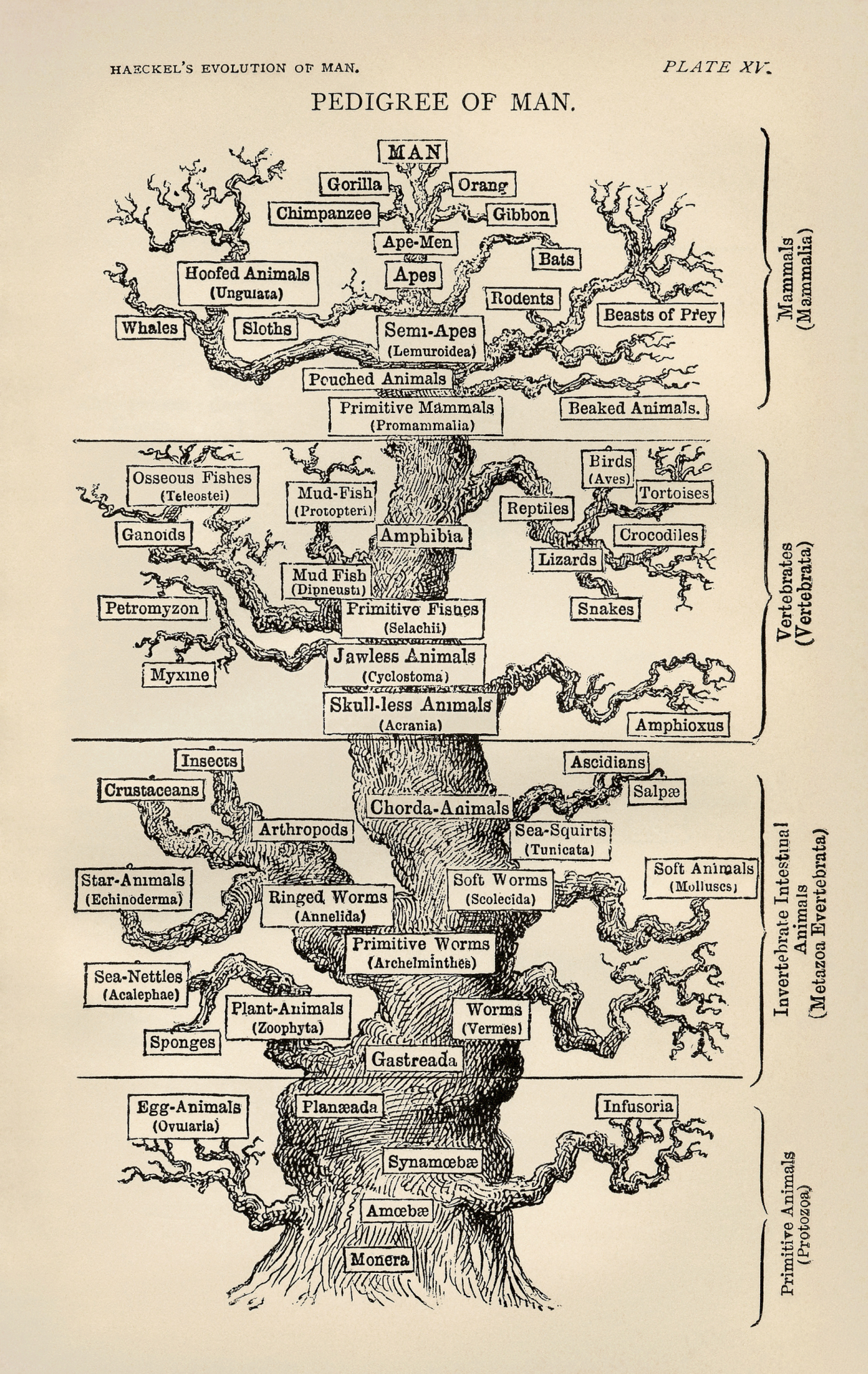
Darwin’s metaphor for the pattern of universal common descent made literal by his German language popularizer Ernst Haeckel in his 1879 The Evolution of Man. “Man” is at the crown of the tree. Wikipedia: Ernst Haeckel
Clade: A group of organisms that consists of a common ancestor and all of its descendants. Biologist Julian Huxley coined the term in 1957. The name of a clade is conventionally plural; the singular refers to each member individually. Valid clades are monophyletic (defined below).
Cladogram: A graphical presentation of the results of phylogenetic/cladistic analyses. They are vaguely tree-shaped (Haeckel’s is explicitly a tree) and all their branches are phylogenetic hypotheses. A cladogram can place the root at the left and branch to the right, or branch from the top down, even place the root at the center and branch in all directions (example below).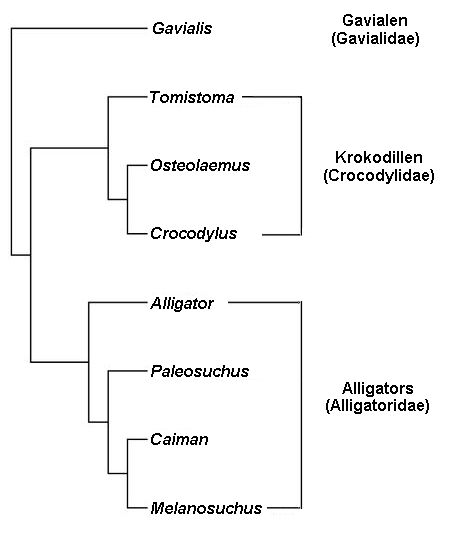
Gavialidae, Crocodylidae and Alligatoridae are clade names here applied to a right-branching phylogenetic tree of crocodylians. As it is missing the common ancestor root at left, it is not a true cladogram. Wikipedia – Clade
Root: The common ancestor of a clade. Except at the root of all life, every root is a branch in an earlier ancestor’s clade, and every common ancestor’s clade of descendants nests within larger clades.
Rootless: Many phylogenetic trees have no root; they may contain many clades but are not a cladogram in their entirety.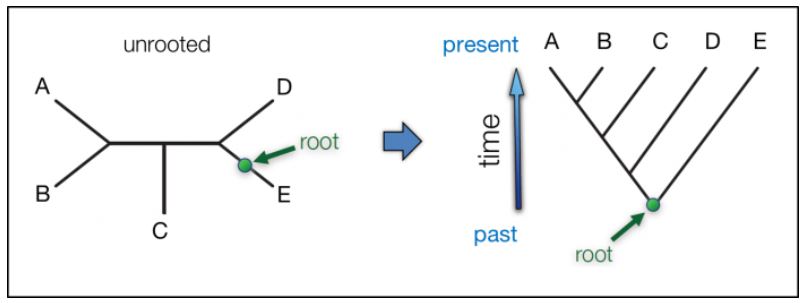
Before and after rooting the phylogenetic tree. (Branch lengths are not to any scale). Many phylogenetic reconstructions don’t estimate root position as that increases the number of possible trees and the time it takes to calculate the tree. Visualize the left tree as made from string; push a pin into the root spot, then rotate the remaining branches around the pin-point to yield the tree on the right. The arrow indicates the direction of evolution as implied by the root position. EMBL-EBI – Intro to Phylogenetics
Taxon (plural taxa): A group of one or more populations of an organism or organisms seen by taxonomists to form a unit. Taxonomists now try to avoid naming taxa that are not demonstrably monophyletic (defined farther below).
Taxonomic Rank: The level of a taxon within a taxonomic hierarchy. Phylum is higher rank than Class, Genus is lower than Family, higher is more general, lower is more specific. To identify a particular organism only genus and species are necessary, specification of higher ranks is not necessary.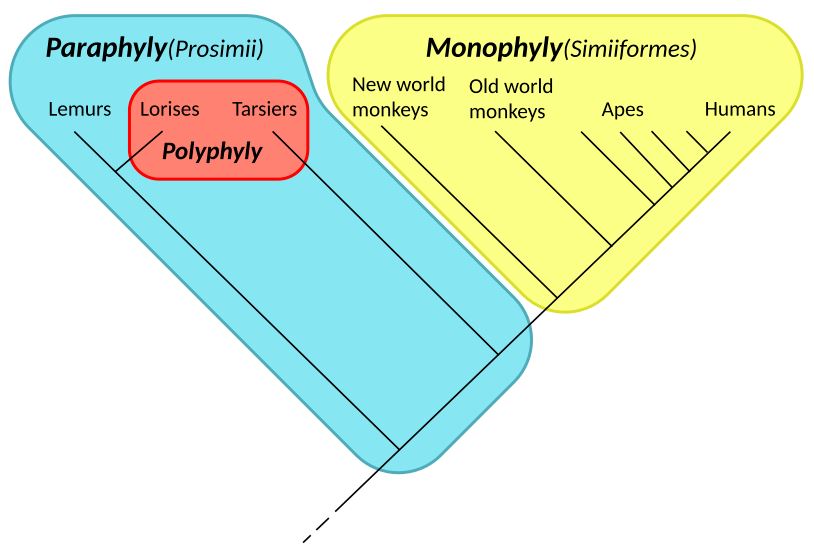
Primate phylogenetic tree (root is located but not identified): Monophyly (yellow) in Simians; Paraphyly in Prosimians (blue, including red); Polyphyly (red) in Lorises & Tarsiers, both night-active primates. Diagram: Chiswick Chap. Wikipedia – Paraphyly
Monophyletic: When a group includes the common ancestor and all of its descendant groups.
Paraphyletic: When a group includes the common ancestor but not all the descendant groups.
Polyphyletic: When a group is derived from more than one common evolutionary ancestor or ancestral group and therefore not truly suitable for placing in the same taxon. With the advent of protein and/or DNA analysis and biological clocks, many groups formerly believed to be monophyletic were revealed to be polyphyletic and thus needed to be disassembled and reclassified into monophyletic (or at least paraphyletic) clades. This goal is not yet fully achieved but a lot of progress has been made. If you’ve noticed that a lot of bird families and/or orders have been disassembled within the past fifteen years with species sent in various directions and new ones inserted, this is why.
Sister taxa, polytomy and basal taxon all in one clade. Quora – Basal
Branch: The branch shows the path of transmission of genetic information from one generation to the next. When branch lengths indicate genetic change or elapsed time, a reference scale or explanation should be supplied; i.e. the longer the branch, the more elapsed time or genetic change (divergence) has occurred. Branch length often signifies nothing beyond appearance for appearance sake.
Branch Tip: The end (tip) of a branch in a phylogenetic tree represents the latest descendant, whether extant or extinct, of that particular lineage.
Node: The points in a cladogram where lines intersect or branch. This point is the common ancestor of both lineages and represents divergences (speciation events).
Nested: When a clade located is located within another clade. In the diagram below, the Great Ape clade nests within the Ape clade which nests within the Catarrhine clade which nests within the Simian clade and so on down the line.
Sister Clades (or taxa): When two clades have an immediate common ancestor. In the diagram below, lemurs and lorises are sister clades, while humans and tarsiers are not.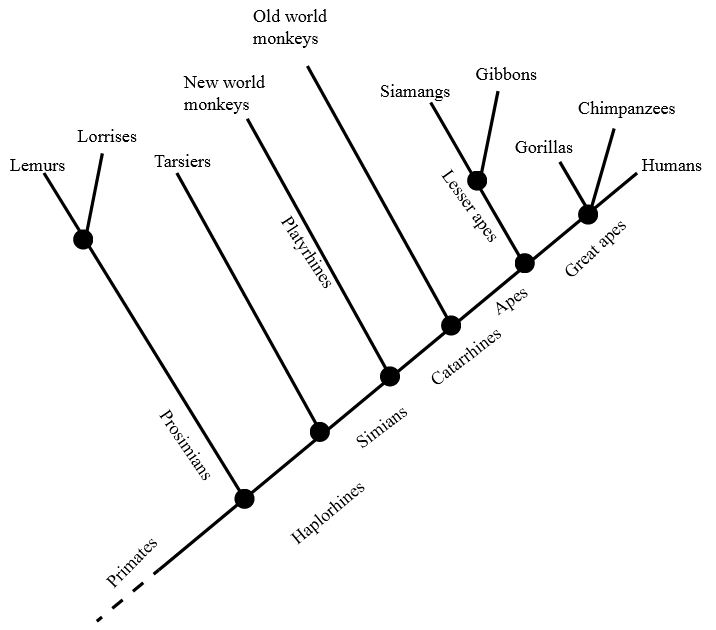
Cladogram of modern primate groups, showing branches, (Prosimians, etc.) branch tips (Tarsiers, etc.), and nodes (black dots where branches diverge (or meet). Above each node is the group name , which could be replaced or augmented by the synapomorphy (common character) as shown in the following two cladograms. All tarsiers are haplorhines, but not all haplorhines are tarsiers; all apes are catarrhines, but not all catarrhines are apes; etc. Wikipedia – Clade
Basal: Close to the root of a tree; the group which gave rise to later forms, a non-judgmental term to replace ‘primitive’ or ‘ancestral.’ A group of organisms which diverge early in the evolutionary history of a clade, forming an ‘outgroup’ to the rest of the clade. Basal forms often retain traits of the ancestral form lost in other and later (‘derived’) descendants, thus providing insights into the early stages of evolutionary history of the clade. Basal traits are original condition of the common ancestor. ‘Advanced’ means the character evolved within a later subgroup of the clade, with no judgment about complexity, superiority or adaptiveness of that trait. In the primate diagram above, the prosimian clade is basal to the haplorhines clade, and both prosimian and tarsier clades are basal to the simian clade. Also see the basal taxon in the second diagram above.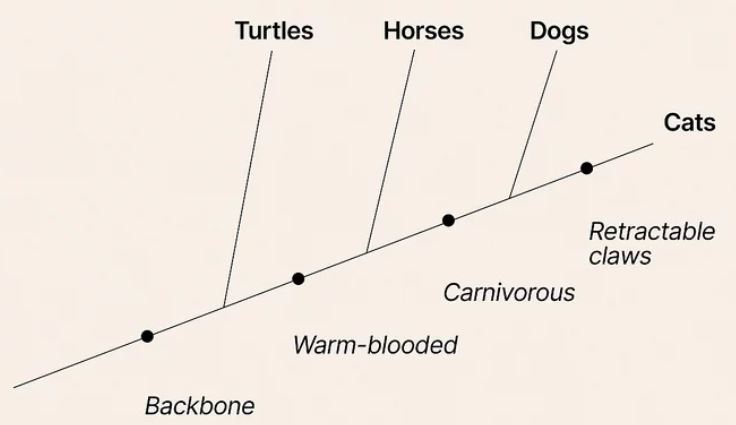
Constructing a simple cladogram using synapomorphies (carnivorous, etc.) rather than group names. Wikihow – Read a Cladogram
Extant: Still existing; the taxon still has living members.
Extinct: No longer existing; no members of the taxon are alive today.
Stem Group: An extinct species or collection of species that is more closely related to an extant clade than the extant clade is to its most closely related sister clade. Example: Modern humans are more closely related to the stem group of extinct hominins (Homo erectus, etc.) than they are to extant chimpanzees.
Crown Group: For a clade to be a crown group it must have extant species with common character(s) (synapomorphy). It will also include all extinct clade members as well as the most recent common ancestor of all clade members whether they are extant or extinct.
Apomorphy: A derived trait; a novel character or character state that has evolved from its plesiomorphy (ancestral form).
Synapomorphy: A character or trait that is shared by two or more taxonomic groups and is derived through evolution from a common ancestral form. The cladogram below designates synapomorphies with red bars. Diagonal trees such as this are preferred when indicating synapomorphies.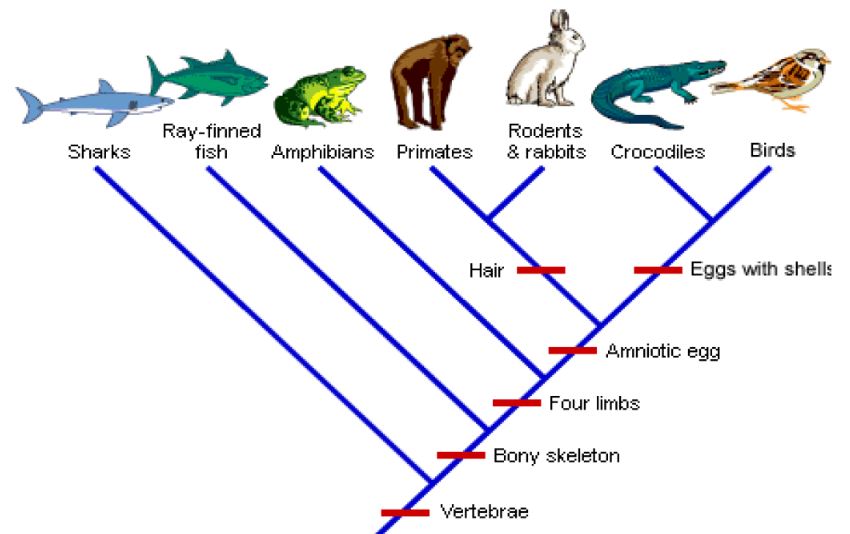
Sample teaching cladogram from: University of British Columbia
Lineage: A single line of descent or linear chain within a tree connecting the common ancestor to a particular tip taxon.
Polytomy: On a phylogeny tree where more than two branches (lineages) descend from a single ancestral lineage. The presumed reason is that we do not yet have enough evidence to strongly support one set of possible relations over other possible sets. It might mean (but probably doesn’t) that we think that the descendant lineages speciated simultaneously. Ranks and individuals within polytomies are frequently labeled incertae sedis, “of uncertain placement.” Complete information should eventually resolve all polytomies.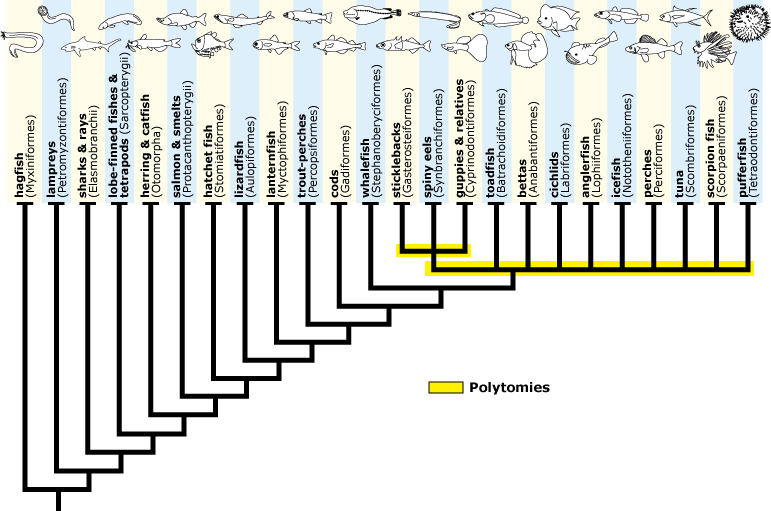
Fish cladogram with two polytomies. UC Berkeley
In the following five postings in this series, many of these terms will appear, especially the term “clade.” Unlike “Superorder, Order, Suborder…”, “clade” gives no indication of where in a taxonomic sequence it falls, and only by identifying the prior and following ranks will one have a clue. To clarify this situation somewhat, where appropriate I’ve inserted the term Cladesubscript 1,2,3…
A term like “Clade19 Class Amphibia” will then indicate a rank 19th in the sequence beginning with Clade1 Domain Eukaryota, as well as the Linnaean rank of Class. This is my own invention which you may find useful, confusing or unnecessary. I’m not entirely happy with it, but if nothing else, it shines a little light on how confusing this nomenclatural problem can get. It also helps when constructing and reading long indented phylogenetic trees and lineages which otherwise can become confusing.
Cladograms can get quite complex. The following pair of cladograms pertain to the virus SARS-CoV-2. Pressing <control>+ makes the diagram larger and (one hopes) clearer.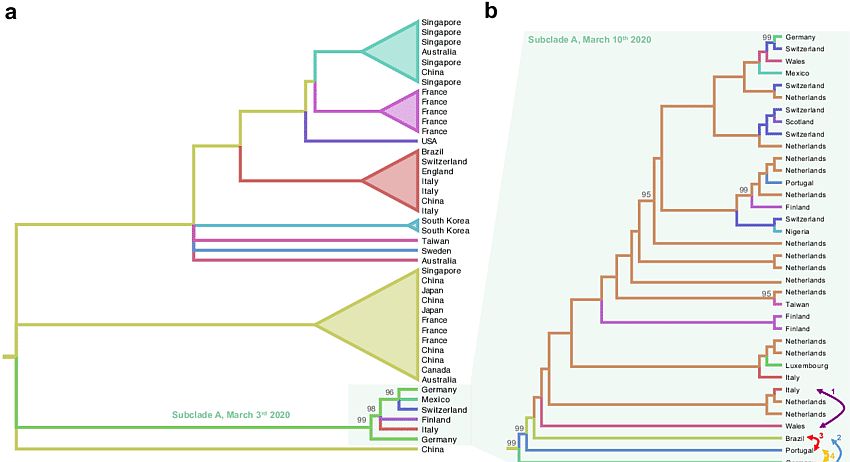
Cladograms of SARS-CoV-2 subclades. Cladograms extracted from ML phylogenies rooted by enforcing a molecular clock. Colored branches represent country of origin of sampled sequences (tip branches) and ancestral lineages (internal branches). Numbers at nodes indicate ultrafast bootstrap (BB) support (only >90% values are shown).
(a) Cladogram of a monophyletic clade within the SARS-CoV-2 ML tree inferred from sequences available on March 3 rd 2020 (Supplementary Figure S1). The subclade including sequences from Italy and Germany, named Subclade A (lower right), is highlighted.
(b) Cladogram of subclade A of the SARS-CoV-2 ML tree including additional sequences available on March 10th 2020 (Supplementary Figure S2).
Link to Research Gate report.
A cladogram of SARS-CoV-2 genetic variants. This may actually be a phylogenetic tree, not a cladogram, as there may be no root defined at the center (but who can tell?).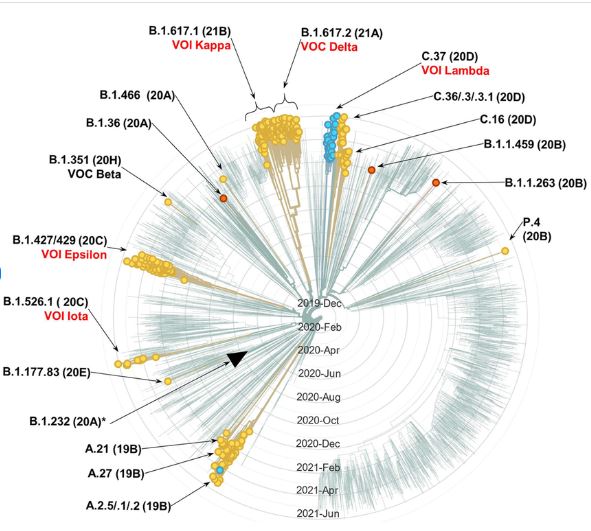 Nextstrain-generated radial cladogram of SARS-CoV-2 genetic variants (as of 12 June 2021). Showing 519 of 3,883 genomes randomly sampled between Jun 2020 and May 2021, belonging to 23 PANGO lineages. Circles tip branches containing genomes with any mutation in L452 (circles on branches without the mutations were manually removed for a visual clarity). Yellow circles, branches with L452R; blue, with L452Q; orange, with L452M. Black triangle tips the branch of lineage that includes CAL.20A. (At the date of analysis, CAL.20A strains were not among the sampled genomes in the Nextstrain database, and only three B.1.232 [all non-CAL.20A] lineages were found). Nomenclature: PANGO lineage followed by Nextstrain clade in parenthesis and the current VOC/VOI designation. In red, VOC/VOI with an omnipresent or nearly omnipresent L452 mutation (VOC beta is in black, as the L452 mutation is found only in few genomes of the variant). Link to Research Gate report.
Nextstrain-generated radial cladogram of SARS-CoV-2 genetic variants (as of 12 June 2021). Showing 519 of 3,883 genomes randomly sampled between Jun 2020 and May 2021, belonging to 23 PANGO lineages. Circles tip branches containing genomes with any mutation in L452 (circles on branches without the mutations were manually removed for a visual clarity). Yellow circles, branches with L452R; blue, with L452Q; orange, with L452M. Black triangle tips the branch of lineage that includes CAL.20A. (At the date of analysis, CAL.20A strains were not among the sampled genomes in the Nextstrain database, and only three B.1.232 [all non-CAL.20A] lineages were found). Nomenclature: PANGO lineage followed by Nextstrain clade in parenthesis and the current VOC/VOI designation. In red, VOC/VOI with an omnipresent or nearly omnipresent L452 mutation (VOC beta is in black, as the L452 mutation is found only in few genomes of the variant). Link to Research Gate report.
When examined close up, the gray area fringing the circular cladogram above is revealed to be tiny cladograms. I can’t imagine how much you’d have to blow it up (and somehow maintain clarity) to be able to actually read it.
From: Research Gate report.
For additional information:
AmphibiaWeb – Taxonomy
The Burgess Shale – Stem groups & crown groups
Digital Atlas of Ancient Life – Crowns & stems
Wikipedia – Clade
Wikipedia – Crown Group
Wikipedia – List of Latin and Greek words commonly used in systematic names
Wikipedia – Phylogenetic nomenclature
The Taxonomy Series
Installments post ever other day; installments will not open until posted.
Taxonomy One: A brief survey of the history and wherefores of taxonomy: Aristotle, Linnaeus and his binomial system of nomenclature, taxonomic ranks and the discovery and application of biological clocks.
Taxonomy Two: Introduces the higher levels of current taxonomy: the three Domains and the four Kingdoms. We briefly discuss Kingdom Protista, then the seven phyla of Kingdom Fungi.
Taxonomy Three: Kingdom Plantae.
Taxonomy Four: Kingdom Animalia to Phylum Annelida.
Taxonomy Five: A discussion of Cladistics, how it works and why it is becoming ever more important.
Taxonomy Six: Phylum Chordata, stopping at Class Mammalia.
Taxonomy Seven: Class Mammalia.
Taxonomy Eight: Class Aves, beginning with a comparison of five different avian checklists of the past 50 years.
Taxonomy Nine: A cladogram and discussion of Subclass Neornithes (modern birds) of the past 110 million years, reaching down to the current forty-one orders of birds.
Taxonomy Ten: A checklist of Neornithes including all ranks and clades down to the rank of the current 251 families of birds (plus a few probable new arrivals) with totals of the current 11,017 species of birds.
Discover more from SANTA MONICA BAY AUDUBON SOCIETY BLOG
Subscribe to get the latest posts sent to your email.


